Introduction
Overview
Teaching: 10 min
Exercises: 0 minQuestions
What is CWL?
What is the goal of this training?
Objectives
Gain a high level understanding of the example analysis.
This lesson has migrated to https://doc.arvados.org/rnaseq-cwl-training/01-introduction/index.html
Introduction to Common Worklow Language
The Common Workflow Language (CWL) is an open standard for describing automated, batch data analysis workflows. Unlike many programming languages, CWL is a declarative language. This means it describes what should happen, but not how it should happen. This enables workflows written in CWL to be portable and scalable across a variety of software and hardware environments, from workstations to cluster, cloud, and high performance computing (HPC) environments. As a standard with multiple implementations, CWL is particularly well suited for research collaboration, publishing, and high-throughput production data analysis.
Introduction to the example analysis
This training uses a bioinformatics RNA-seq analysis as a motivating example. However, specific knowledge of the biology of RNA-seq is not required for these lessons. For those unfamiliar with RNA-seq, it is the process of sequencing RNA present in a biological sample. From the sequence reads, we want to measure the relative numbers of different RNA molecules appearing in the sample that were produced by particular genes. This analysis is called “differential gene expression”.
The entire process looks like this:
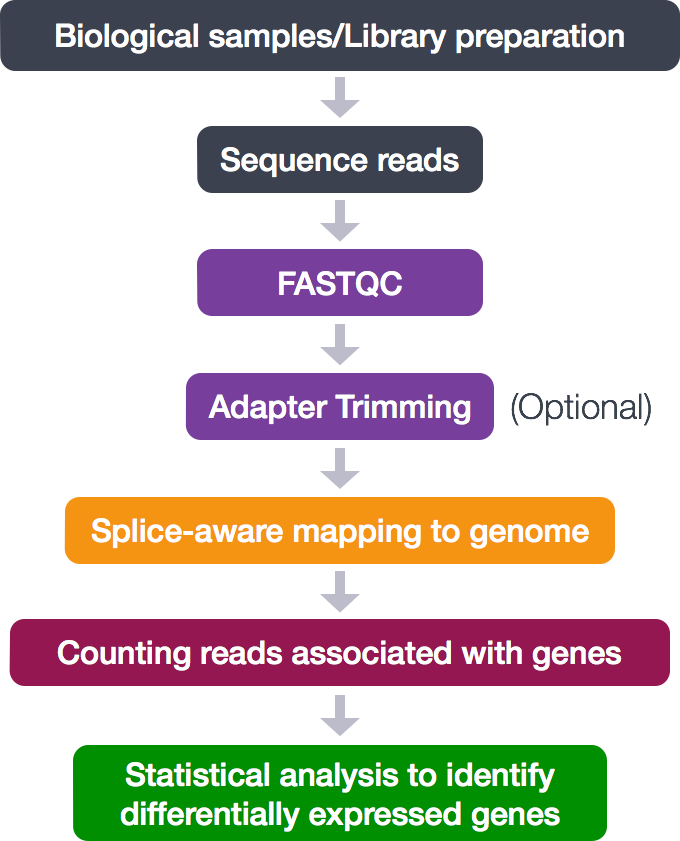
For this training, we are only concerned with the middle analytical steps (skipping adapter trimming).
- Quality control (FASTQC)
- Alignment (mapping)
- Counting reads associated with genes
In this training, we do not develop the analysis from first principals, instead we we will be starting from an analysis already written as a shell script, which will be presented in lesson 2.
Key Points
Common Workflow Language is a standard for describing data analysis workflows
We will use an bioinformatics RNA-seq analysis as an example workflow, but does not require in-depth knowledge of biology.
After completing this training, you should be able to begin writing workflows for your own analysis, and know where to learn more.